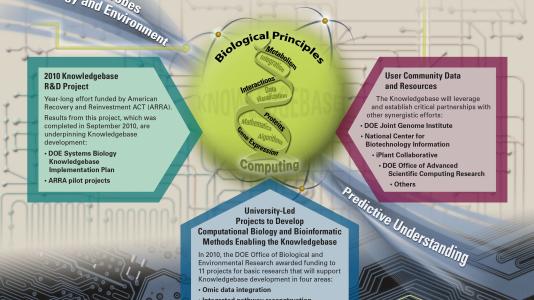
The goal of this collaboration is to develop a Systems Biology Knowledgebase (KBase), designed to accelerate our understanding of microbes, microbial communities and plants.
Microbes are the oldest form of life on earth—and vital to life as we know it. Without these single-cell organisms we would not, for example, have an atmosphere that supports human life or digest food efficiently. The more recently evolved and complex plants are also central to our lives.
Microbes and plants harvest energy from the sun and drive environmental cycles. DOE is discovering how the powerful capabilities of these organisms and their communities can help solve problems in energy and environmental balance and remediation. The basic knowledge gained from studies of these organisms impacts all areas of agriculture and extends into animal and human health.
In order to address many of these problems, however, researchers must analyze mountains of data, spend considerable time integrating data, or develop analytical tools. But with consistent and rapid improvements in technology, scientists are no longer able to keep pace with the rate of data generation, which includes interrelated data of different types. KBase will provide enhanced power and integration capabilities to make rapid comparisons across multiple genomes and organisms through free and open access to these data, models and simulations.
KBase will be a community-driven, extensible and scalable software framework and application system. As a distributed system, it will leverage major existing scientific capabilities and supercomputing facilities within the DOE national laboratory complex. Adam Arkin, the KBase team’s lead principal investigator (PI), from Lawrence Berkeley National Laboratory, describes KBase as, “More than a database. It will also be a powerful modeling framework that transforms data about microbes, plants and their communities into models of their function. It will be an open system allowing the community to integrate their data and compuational tools more easily and thereby create a ‘network effect’ among researchers in biological systems science.”
“The goal is to enable researchers to both design and interpret their experiments more powerfully, and to share those results and their interpretations,” said Rick Stevens, co-PI and associate laboratory director for Computing, Environment and Life Sciences at Argonne. “New planned functionality will allow users to visualize data, create models or design experiments based on KBase suggestions.”
KBase will be composed of a series of core biological analysis and modeling functions, including an application programming interface (API) which can be used to interface between different software programs within the community. These capabilities will be constructed from the popular analysis systems at each of the KBase sites, such as Argonne’s MG-RAST and SEED systems. Their integration into KBase will combine their individual functions to create the next generation of biological models and analysis tools.
The KBase API will enable third-party researchers to design new functions as well. “KBASE will enable researchers to work with next generation sequencing data without the need to be some sort of renaissance scientist understanding computing, next-generation sequencing, statistics, computational biology and molecular ecology concepts,” said Folker Meyer, computational biologist at Argonne.
KBase will be supported by a computing infrastructure based on the OpenStack cloud system software, distributed across the core sites. These resources will initially be based on existing hardware at each of these sites. As computing demands grow, the team will evaluate commercial cloud options as well.
A cloud-style project, KBase will make biological data and analysis capabilities available behind a series of well-structured APIs that will be accessible by remote users. These abstractions will allow the team to do the required engineering work behind the scenes to scale the computing infrastructure and services that provide these APIs.
Several universities and DOE national laboratories are partnering to create the first implementation of this comprehensive platform. The collaboration is led by Lawrence Berkeley National Laboratory and includes participation from Argonne, Brookhaven and Oak Ridge. Also participating the multi-institutional program are the the Cold Spring Harbor Laboratory; University of California, Davis; Hope College in Michigan; the University of Illinois at Urbana-Champaign; and Yale University. The Joint Genome Institute and several university knowledgebase projects are also identified as important contributors.
The project is funded by DOE’s Genomic Science program, which funds research aimed at identifying the fundamental principles that drive biological systems relevant to DOE missions in energy and the environment.
For more information on this program, please visit http://genomicscience.energy.gov/index.shtml