A team of researchers from Argonne National Laboratory and the University of Chicago has received a best paper award at Supercomputing 2016 in Salt Lake City, Utah. SC is the premier international conference on high-performance computing, networking, storage, and analysis. The paper was presented November 13, 2016, at the SC workshop “Computational Approaches for Cancer at SC16.”
The winning paper – co-authored by Argonne researchers Jonathan Ozik, Nicholson Collier, Justin M. Wozniak, and Charles Macal and University of Chicago researchers Chase Cockrell, Melinda Stack and Gary An – describes the use of a high-performance agent-based model to explore mutation patterns in simulations of colorectal cancer.
Agent-based models integrate multiple scales of behavior and are increasingly being used in cancer research. Many model executions are typically needed to explore various parameter setting and experimental conditions. Hence, adaptive ensemble-based model exploration techniques are often used; but the approach has the disadvantage of being constrained to a small fraction of the parameter space, primarily because of the high computational cost in carrying out the necessary model evaluations.
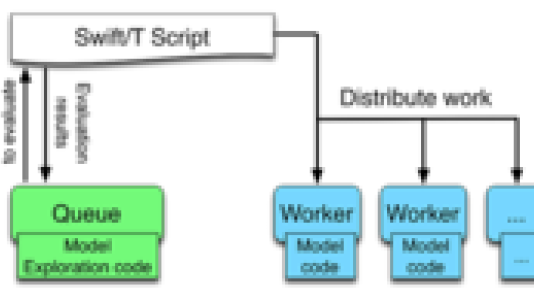
To increase the potential benefits of agent-based modeling and other complex modeling approaches, the Argonne/University of Chicago team devised a new framework, called Extreme-scale Model Exploration with Swift (EMEWS), that takes advantage of high-performance computing to run large, highly concurrent ensembles of simulations of varying types, while supporting a wide class of model exploration algorithms.
Key to the experiments was the use of Swift, a parallel programming system developed at Argonne, and Mira, an IBM Blue Gene/Q supercomputer at the Argonne Leadership Computing Facility. The researchers configured EMEWS to fit model outputs with epidemiological data provided by the National Cancer Institute. They then ran an ensemble of simulations on Mira, each involving an agent-based model of cancer cells including their DNA mutations and resultant loss of functions.
“The ensembles sought to extract an unknown biomedical phenomenon – the ability of cells to repair DNA damage,” said Justin Wozniak, a computer scientist in Argonne’s Mathematics and Computer Science Division. “Mira enabled us to apply many simulation results to hone in on the answer, and Swift made it easy to deploy at large scale. Our largest Swift run conducted 6 million simulations on 64,000 cores of Mira, delivering the equivalent of 1001 CPU-days in just 22 minutes, or 4,570 simulations per second.”
The researchers won an NVIDIA Titan X graphics card, which they plan to use in prototyping future work. The objective will be to determine whether accelerator systems can unlock even greater amounts of parallelism needed to tackle the computational requirements of agent-based models of biological systems.
The paper is available on the web: http://www.mcs.anl.gov/~wozniak/papers/Cancer2_2016.pdf
Information about the workshop is on the SC website: http://www.scworkshops.net/cancer2016